Tessa Cook
RSNA Large Language Model Benchmark Dataset for Chest Radiographs of Cardiothoracic Disease: Radiologist Evaluation and Validation Enhanced by AI Labels (REVEAL-CXR)
Jan 21, 2026Abstract:Multimodal large language models have demonstrated comparable performance to that of radiology trainees on multiple-choice board-style exams. However, to develop clinically useful multimodal LLM tools, high-quality benchmarks curated by domain experts are essential. To curate released and holdout datasets of 100 chest radiographic studies each and propose an artificial intelligence (AI)-assisted expert labeling procedure to allow radiologists to label studies more efficiently. A total of 13,735 deidentified chest radiographs and their corresponding reports from the MIDRC were used. GPT-4o extracted abnormal findings from the reports, which were then mapped to 12 benchmark labels with a locally hosted LLM (Phi-4-Reasoning). From these studies, 1,000 were sampled on the basis of the AI-suggested benchmark labels for expert review; the sampling algorithm ensured that the selected studies were clinically relevant and captured a range of difficulty levels. Seventeen chest radiologists participated, and they marked "Agree all", "Agree mostly" or "Disagree" to indicate their assessment of the correctness of the LLM suggested labels. Each chest radiograph was evaluated by three experts. Of these, at least two radiologists selected "Agree All" for 381 radiographs. From this set, 200 were selected, prioritizing those with less common or multiple finding labels, and divided into 100 released radiographs and 100 reserved as the holdout dataset. The holdout dataset is used exclusively by RSNA to independently evaluate different models. A benchmark of 200 chest radiographic studies with 12 benchmark labels was created and made publicly available https://imaging.rsna.org, with each chest radiograph verified by three radiologists. In addition, an AI-assisted labeling procedure was developed to help radiologists label at scale, minimize unnecessary omissions, and support a semicollaborative environment.
Improving the Performance of Radiology Report De-identification with Large-Scale Training and Benchmarking Against Cloud Vendor Methods
Nov 06, 2025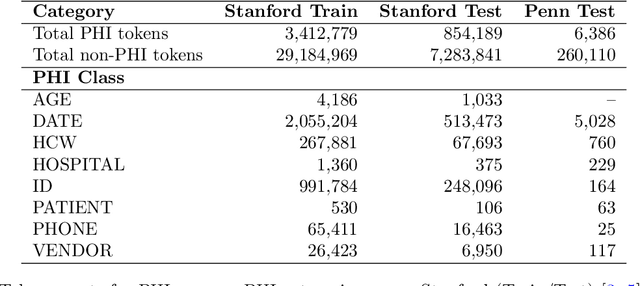

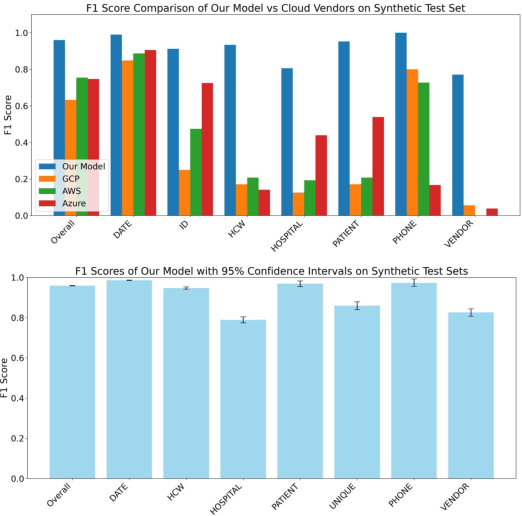
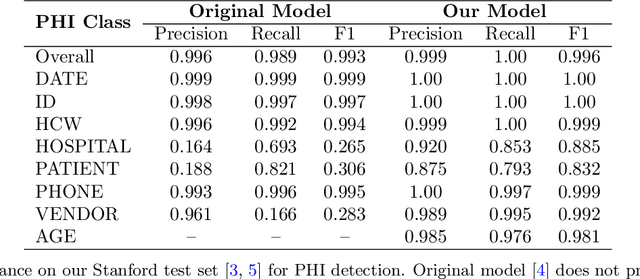
Abstract:Objective: To enhance automated de-identification of radiology reports by scaling transformer-based models through extensive training datasets and benchmarking performance against commercial cloud vendor systems for protected health information (PHI) detection. Materials and Methods: In this retrospective study, we built upon a state-of-the-art, transformer-based, PHI de-identification pipeline by fine-tuning on two large annotated radiology corpora from Stanford University, encompassing chest X-ray, chest CT, abdomen/pelvis CT, and brain MR reports and introducing an additional PHI category (AGE) into the architecture. Model performance was evaluated on test sets from Stanford and the University of Pennsylvania (Penn) for token-level PHI detection. We further assessed (1) the stability of synthetic PHI generation using a "hide-in-plain-sight" method and (2) performance against commercial systems. Precision, recall, and F1 scores were computed across all PHI categories. Results: Our model achieved overall F1 scores of 0.973 on the Penn dataset and 0.996 on the Stanford dataset, outperforming or maintaining the previous state-of-the-art model performance. Synthetic PHI evaluation showed consistent detectability (overall F1: 0.959 [0.958-0.960]) across 50 independently de-identified Penn datasets. Our model outperformed all vendor systems on synthetic Penn reports (overall F1: 0.960 vs. 0.632-0.754). Discussion: Large-scale, multimodal training improved cross-institutional generalization and robustness. Synthetic PHI generation preserved data utility while ensuring privacy. Conclusion: A transformer-based de-identification model trained on diverse radiology datasets outperforms prior academic and commercial systems in PHI detection and establishes a new benchmark for secure clinical text processing.
Current State of Community-Driven Radiological AI Deployment in Medical Imaging
Dec 29, 2022



Abstract:Artificial Intelligence (AI) has become commonplace to solve routine everyday tasks. Because of the exponential growth in medical imaging data volume and complexity, the workload on radiologists is steadily increasing. We project that the gap between the number of imaging exams and the number of expert radiologist readers required to cover this increase will continue to expand, consequently introducing a demand for AI-based tools that improve the efficiency with which radiologists can comfortably interpret these exams. AI has been shown to improve efficiency in medical-image generation, processing, and interpretation, and a variety of such AI models have been developed across research labs worldwide. However, very few of these, if any, find their way into routine clinical use, a discrepancy that reflects the divide between AI research and successful AI translation. To address the barrier to clinical deployment, we have formed MONAI Consortium, an open-source community which is building standards for AI deployment in healthcare institutions, and developing tools and infrastructure to facilitate their implementation. This report represents several years of weekly discussions and hands-on problem solving experience by groups of industry experts and clinicians in the MONAI Consortium. We identify barriers between AI-model development in research labs and subsequent clinical deployment and propose solutions. Our report provides guidance on processes which take an imaging AI model from development to clinical implementation in a healthcare institution. We discuss various AI integration points in a clinical Radiology workflow. We also present a taxonomy of Radiology AI use-cases. Through this report, we intend to educate the stakeholders in healthcare and AI (AI researchers, radiologists, imaging informaticists, and regulators) about cross-disciplinary challenges and possible solutions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge